Recently, Euretos was invited to speak at the Lorentz Conference on ‘How to Make Data FAIR for Open Science’. The conference was organised by the universities of Barcelona, Leiden, Madrid and Vitória (Brazil) and sponsored by Nature Genetics, the European research infrastructure Elixir and Dutch research organisations DTL and BioSB. The conference was recently discussed in an Editorial in Nature Genetics
The aim of the conference was to help researchers from various disciplines publish their research data in an interoperable ‘FAIR’ format: Findable, Accessible, Interoperable and Reusable. During the 4 days workshop researchers, bioinformaticians and data experts worked hand in hand to transform their data into FAIR data, which was the end goal of the workshop.
Euretos was invited to provide a demonstrator of how FAIR data can be used to add value to the individual researcher. FAIR data is not an end in itself, the purpose in the end is to enhance the scientific research process. Euretos has been a participant in the FAIR data movement from its inception and one of the co-authors on the on landmark Nature article on this topic.
The Euretos AI Platform integrates over 200 data sources in its platform and, as such, demonstrates the value of integratable and reusable data. The more FAIR data is, the easier it is for computers to integrate it and derive additional value from it. A key topic was how to deal with the enormous amount of data that a research is confronted with if hundreds of data sources are combined.
Take for example the 175+ life sciences data sources that have been integrated in the Euretos Knowledge Platform. It is of course great to have all this data together in one single view. On the other hand it also makes painfully clear the amount of data that is available for a researcher to assess:
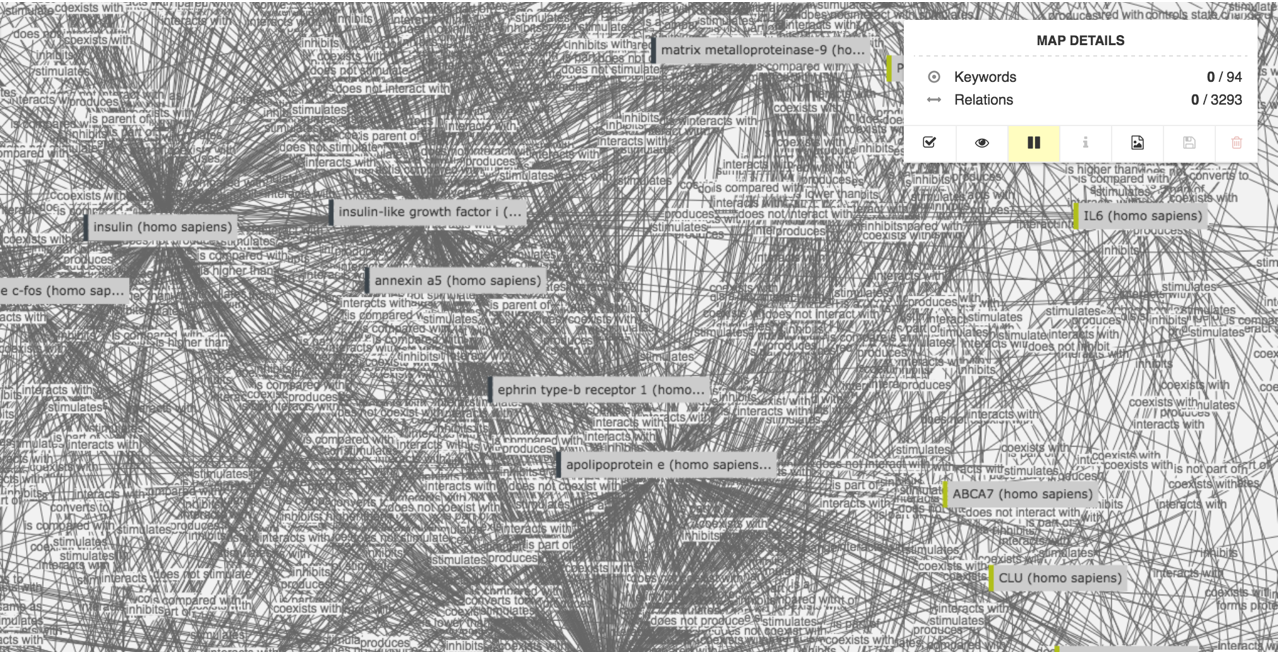
In the above example, 94 genes associated with presenile dementia have amongst themselves close to 3300 relations. And this only covers the genetic interactions!
These ‘ridiculograms’ are clearly not human-interpretable and require smart ways to deal with the data overload. For us this data overload has been a major challenge and to address it we use the parallel of the hourglass:

Basically you first need approaches to bring down the number of potential candidates to a relevant short list: go from a high volume of candidates to just a few. Then you can expand the detail again and add relevant key ‘multi-omics’ players interacting with the short-list. This way you can move from high volume to high detail in a controlled manner.
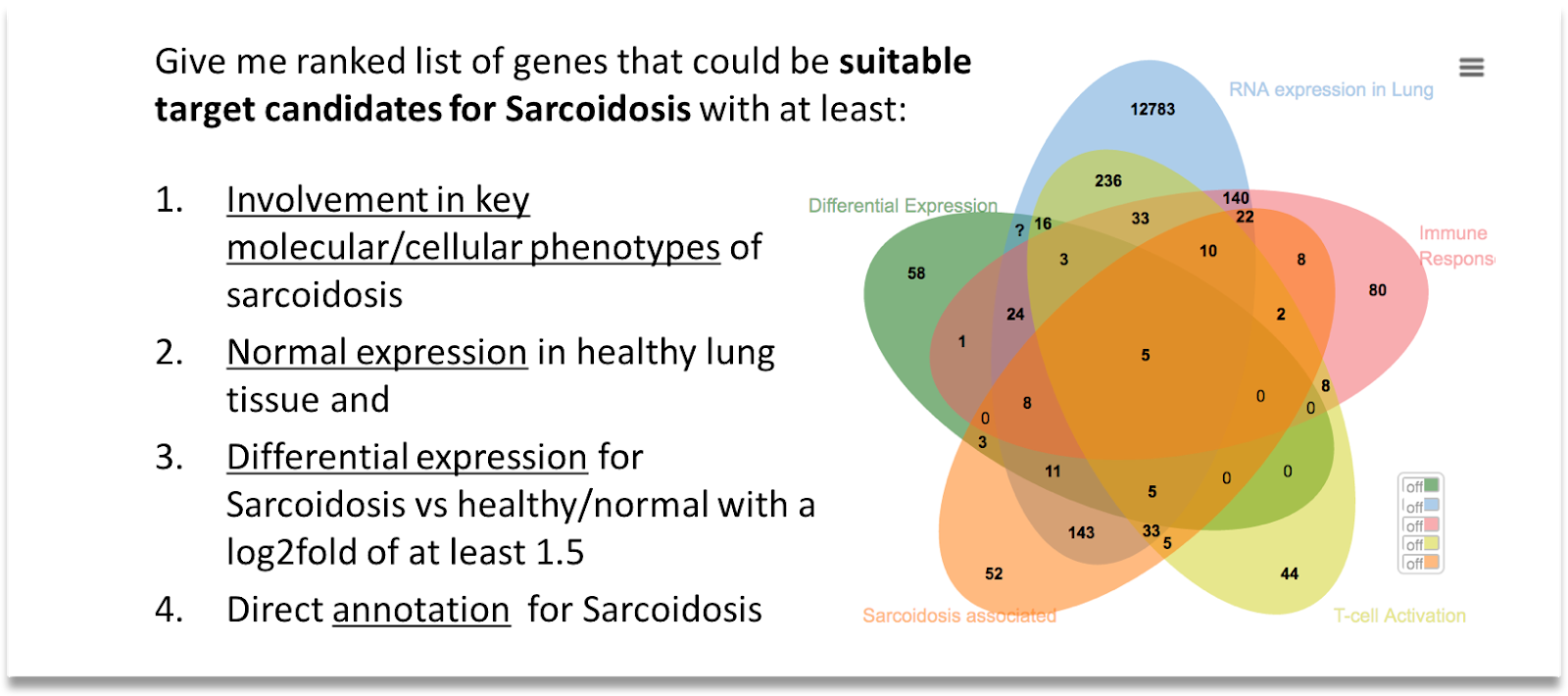
An example of how we enable the researcher to manage the volume of results is intersecting. In this approach the user is able to create and overlay sets of concepts (genes, metabolites, pathways etc.), each representing specific criteria as shown in the Sarcoidosis example below
In addition to this approach other strategies are also possible such as using ranking, sorting and scoring algorithms.Having reached a much more manageable set of concepts, the researcher can now start to expand the analysis and add further detail by identifying key players such as such as pathways, cell & tissue functions, interacting genes, proteins and small molecules.
FAIR data therefore provides new challenges, especially in the area of the volume of data that the researcher needs to manage. This challenge is daunting, but with the appropriate tools to navigate high volume as well as high complexity, significant progress can be made
Subscribe to our mailing list and receive our quarterly updates!
Read our Terms and Conditions

Keep up with our latest news and events. Sign up for our newsletter.